Bio-inspired PCR for Planetary Scale Microbial Discovery
How a genetic quirk in bacteria could help us map the microbial world

Disclaimer: Phage biology is a rapidly evolving field. All the data and literature I present in this piece are current as of May 14th, 2025. If you’re reading from the far future, I always recommend checking the latest primary research.
Discovering microbes isn’t some arcane pastime—it’s the heart of microbiology.
Flip through our scientific timeline, and you’ll notice microbes everywhere.
From Iceland to India, yeast have helped humans ferment for thousands of years. Bacteriophages enabled the discovery of CRISPR. Even molds led to the discovery of penicillin, enabling a generation of antibiotics that extended the human lifespan by nearly two decades (Hutchings et al., 2019)
The pattern is strikingly clear. Some of humanity’s most impressive biological triumphs— from vaccines to restriction enzymes—were either enabled by the discovery of a new microbe or inspired by its unique biology.

And yet, as one of the most exciting frontiers in science, microbiology remains shockingly unexplored. In one review, Rappuoli et al. (2023) estimated that our planet harbors at least a trillion unique species of microbes. But scroll through NCBI’s dataset of microbes and you’ll find ~100,000 species — less than .00001% of what exists.
If our scientific empire is built on such a tiny sliver of the microbial world, what possibilities are we missing out on? And more importantly—what’s stopping us from finding more microbes?
As it turns out, there’s a reason for this. It’s less that we don’t want to and more that we’re bottlenecked by our tools.
Today’s methods of isolating microbes are inherently selective. When we hunt for viruses, we select against a host. When we culture bacteria or fungi, we grow them in media that nurture some cells and starve others. If we want to enable the microbial renaissance, we need a truly agnostic test — one that can identify microbes indiscriminately and at scale.
Meanwhile, at the Pasteur Institute, Mart Krupowic’s lab has discovered a novel element in bacterial genomes capable of replicating DNA without primers. While these modules, termed pipolins, have shaken a basic axiom of DNA replication, they’ve largely been sidelined as a scientific curiosity. (After all, why would anyone want a non-specific DNA polymerase?)
And yet, it seems like we’ve overlooked the most interesting application of pipolins — microbial discovery. The same machinery that lets pipolins replicate DNA could enable the world’s first primerless PCR. Effectively, this would let us amplify trace genomes from bacteria, viruses, and parasites — regardless of how little we know of their sequence or structure. It would offer the first complete snapshot of our microbial world.
Our best science has always emerged when a compelling idea finds its proper use. Pipolins are up next.
Hunting for Microbes
There are already many ways to catch microbes in the wild. Most methods either resort to growing microbes from the environment, or directly sequencing their genomes.
However, as I demonstrate in my earlier piece, both systems suffer from the microbial analog of selection bias. Even the simple act of streaking soil samples on a plate filters bacteria—in this case, based on their ability to metabolize nutrients in the media1. Today’s methods of isolating microbes are inherently selective, and therefore exclusive.
On the other hand, techniques like metagenomic sequencing let us probe entire microbial communities, but only when there’s enough genome from each resident. With the current detection thresholds of metagenomic sequencing, we’re likely missing out on trace microbes that make up most of our microbial diversity.

We can amplify microbes with PCR, but only when we have specific primers for their DNA. But this quickly leads to a chicken-and-egg dilemma. To amplify a microbe, we need its sequence. But in order to sequence, the microbe already needs to be amplified.
While techniques like these are handy for isolating microbes with known properties, they’re blind to everything else. But what if we weren’t looking for any microbe in particular? What if we wanted to cast the widest net possible and narrow down later
That would call for a fundamentally different net. And this is what pipolins will let us build.
The Lost World of Pipolins
Long ago, back when biology was buzzing with the discovery of DNA polymerase, biochemist Arthur Kornberg isolated a curious protein called DnaG.
With further experiments, Kornberg showed that DnaG didn’t string together novel DNA, but instead laid down short scaffolds of RNA where DNA polymerase begin replication (Kornberg et al., 1978).
Since then, our knowledge of primers has flourished. In animal cells, primers are built with RNA. In certain viruses, proteins like VpG shoulder this role, latching onto the genome and serving as a foothold for replication to begin.
While the form and process differ, priming itself was always seen as universal. At least, until recently.
In 2017, Mart Krupowic and his group at the Pasteur Institute published a paper that quietly fractured this assumption. While comparing polymerases across microbial clades, the authors identified a novel genetic element. After isolating the resulting enzyme, they found it had the strange ability to replicate unprimed DNA, and decided to call it primer-independent polymerase B (pipolB).
Further exploration revealed this wasn’t a stroke of luck—similar genetic modules were found across the bacterial kingdom, in certain phages, and stowed away in symbiotic organelles like the mitochondria.

To understand this better, I’ll let the authors explain directly:
Collectively, these results indicate that piPolB from E. coli 3-373-03_S1_C2 pipolin is able to initiate and perform DNA replication of circular and linear templates in the absence of pre-existing primers or additional protein factors.
Considering how radical this finding is, it’s a surprise it hasn’t echoed further throughout the scientific community.
When you follow this logic, it opens up an incredible possibility for microbial discovery. With the unique capacity of pipolins to join nucleotides from scratch, we could design a non-specific PCR — one that could amplify all the microbial genomes in a sample. By enabling replication without exact primers, this approach could capture the presence of microbes that otherwise go undetected.
Of course, this makes some major assumptions. Let’s think through them in single file.
Q: But don’t microbiologists already have tools to amplify all the genomes in sample? Isn’t this a relatively common approach?
It is true that certain fields in microbiology (specifically gut microbiology) have methods to replicate genomes from entire microbial communities.
The most common technique, known as whole genome amplification (WGA), relies on massive libraries of random primers in hopes of initiating DNA replication everywhere. It’s the genetic analog of a brute force search.
Mathematically, this doesn’t make much sense. Depending on the genome, primers can range from 18-30 nt. Even assuming the smallest size, a truly comprehensive library would need >60 billion unique primers2. Our largest libraries today range up to 24-mers, meaning we still miss out on microbes since we don’t cover the full space of primers.
Even if it did, combinatoric chemistry is expensive. While WGA excels at one-off tests, it quickly becomes impractical as we scale to thousands of samples — let alone the millions we’ll need to map the planetary microbiome.
This is why pipolins aren’t just a convenience. By doing away with the priciest piece of genome amplification (namely, our giant libraries of primers), pipolins could let us replicate DNA as long as we feed our system with dirt-cheap dNTPs and ATPs. It’s a radically different technique since it can scale to unseen volumes of microbes.
Q: But aren’t most polymerases sequence-specific? Do we have any idea if pipolBs can replicate DNA aside from the sequences they normally act on?'
Fortunately, they do.
In the original paper, Krupovic’s group showed that pipolB could initiate replication across a giant space of sequences — from native DNA in E. coli to fully-synthetic 33-mer strands of deoxythymidine.
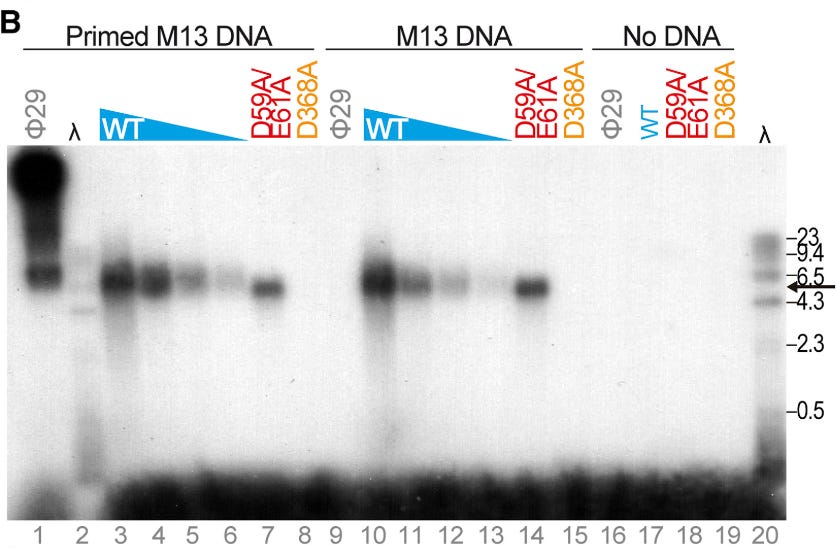
To illustrate, here’s another quote from the paper:
Furthermore, replication of homopolymeric DNA substrates suggests that, contrary to canonical DNA primases (Frick and Richardson, 2001; García-Gómez et al., 2013), piPolB DNA-priming capacity does not rely on a specific template sequence.
For now, it seems that if the target molecule is dsDNA and it’s structurally accessible, pipolB can replicate it.
Though it’s tough to prove that pipolB acts on all DNA, it’s reasonable assume that it operates on an incredibly broad sequence space.
While non-selective polymerase would be a bug in any other context, it’s exactly the feature we need for microbial discovery.
Q: Even if we could use pipolins to replicate mixed populations of DNA, could we ever assemble reads from so many unique genomes?
As enzymes, we expect DNA polymerases to function as long as they operate in the proper environment (i.e. pH, temperature, cofactors), and are constantly replenished with nucleotides.
If anything, the limiting factor is our ability to assemble DNA.
Currently, our best algorithms to assemble reads from metagenomic sequencing (i.e. SCAPP, Meta Flye, etc.) still need coverages of at least 20-fold to function properly (Yu et al. 2023).
Consider a lone bacterium. If, as a crude estimate, we assume a genome of 5Mbp, replicating this 20-fold is already a massive feat3. Now consider a complex microbial community with billions of bacteria. At least intuitively, this seems even more challenging.

Physically, there must be some upper bound to how many microbes we can resolve in a community. In any case, if there’s a bottleneck in this process, it likely won’t be the polymerase, but the number of genomes we can fit in solution and assemble successfully4.
From a purely biological perspective, Krupovic’s paper rebuts a core assumption about DNA replication. And yet, a quick search on Google Scholar shows that this paper received only 34 citations. At least for the time being, the community seems to view pipolins as a scientific curiosity.
But rightly so.
Historically, DNA primases have glimmered because of their specificity. When we first unraveled their function, our most earliest and most impactful solutions used it to selectively amplify DNA. Think about approaches like PCR, which let us identify bacteria, viruses based on particular sequences.
It’s quite natural that the idea of primerless replication hasn’t caught on. What use could anyone possibly have for non-specific DNA replication?
In practically every use case, it makes sense to seek out precise sequences of DNA. But microbial discovery could be the most important exception.
In this singular, yet incredibly meaningful edge case, this quirk of pipolins isn’t a bug, but a feature. When we scour the planet for genetic footprints of unknown microbes, we want to amplify everything indiscriminately.
Perhaps no one reading this paper fancies hunting microbes. Pipolins might be exactly what this field was waiting for.
Revamping the Microbial Discovery Pipeline
As I mentioned earlier, microbes today are largely discovered through brute force. We begin by collecting samples from the environment, isolate microbes using mostly arbitrary selection filters, then sequence our results. While this approach is routinized and reliably churns out new species, fundamentally wrong. What we isolate simply isn’t representative of the world.
Because of their inherent design, we aren’t seeing all microbes in the planet — merely the small fraction that are responsive to our tests.
If we want to resolve the microbial world (and maintain our sanity) we can’t use a piecemeal approach. We need an agnostic system that can identify all microbes in a sample.
It wouldn’t take much to integrate pipolins into microbial discovery. Here’s one protocol I designed from my work isolating viruses.
Consider a soil sample. After suspending the sample in an arbitrary buffer (i.e. SM, PBS, etc.) and filtering through a 0.2 micron membrane, we’ll have a solution packed with viral particles and small molecules. Treating this with a lysis buffer will give us a batch of exposed DNA.
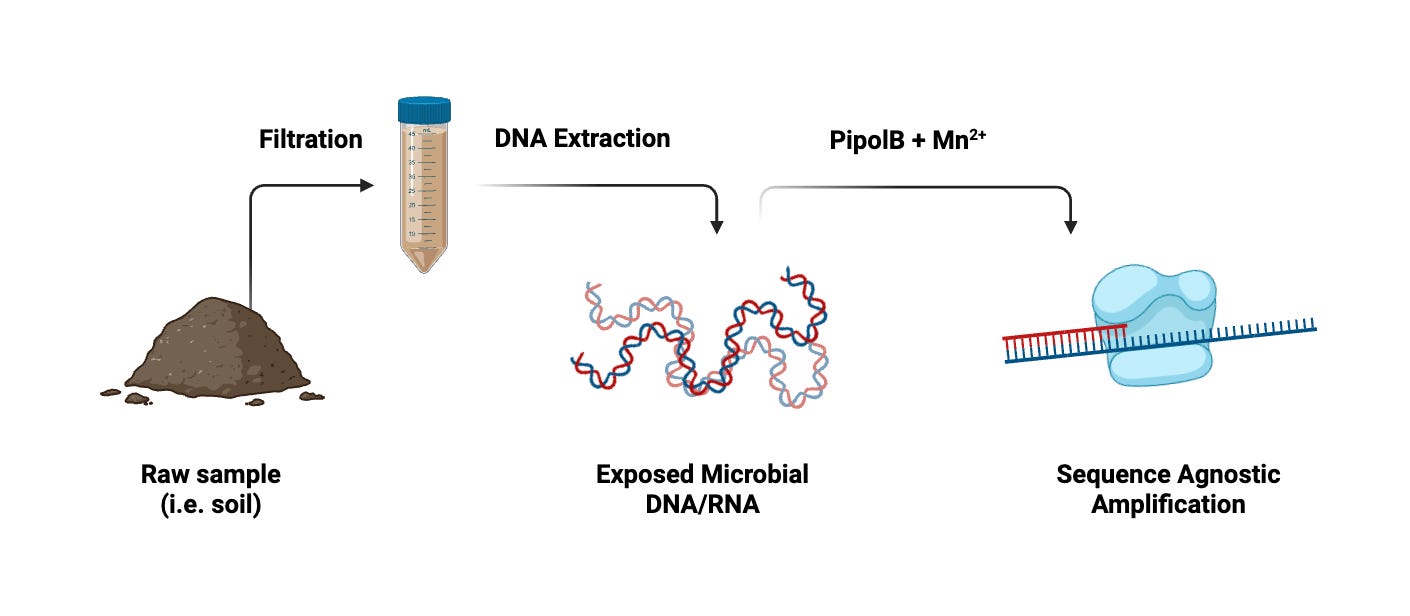
Alternatively, if we wanted to resolve RNA (as is often the case with animal viruses), we would simply use RT to turn the entire sample into DNA first. Then, much like PCR, we would heat the sample to de-anneal DNA and treat it with pipolB to begin the replication process.
Since pipolins are distinct from normal polymerases, they have unique enzymatic needs. While current literature suggests that pipolB functions optimally with manganese (Mn2+), these parameters can always be optimized with concentration testing.
From here, we would follow the standard PCR heating cycle to amplify all the genomes in our sample.
Then, after sequencing with standard techniques (i.e. Illumina, PacBio), we could then resolve our assembled reads.
As you can see, the bulk of the process remains the same. The end result is still snapshot of a microbial community— the only difference is that we now have a panoramic view.
And Now, Your Turn
As a virus hunter, I believe that microbial exploration is one most important scientific pursuits.
If you’re reading this article, assume you care about this, too.
Microbiology has already changed our world. Now we’re nearing a precipice where microbiology becomes our world and makes it larger.
The community of microbiologists is still small, and that gives us the chance to build the tools that will define this field for generations.
Over the next few weeks, I’ll be in touch with the Krupowic lab to see how our ideas can intersect, and potentially running experiments with pipolins on phage DNA.
This article is an open invitation. If you want to help design and test this approach, write to me and we can find ways to collaborate.
Let’s make this real.
Footnotes
And no, this distinction isn’t just wordplay. Some of the most clinically relevant bacteria today (including spirochetes responsible for syphilis) weren’t studied for decades because they couldn’t be grown on traditional growth media. This is the drawback to selective tests — they assume that what we see is all there is.
This would be a conservative estimate since we ran four to the eighteenth power, which models the combinatorics of our shortest primers.
Here, I use the term “bacterial genome” to illustrate the downstream effects of continuing DNA replication. For viruses, fungi, protists, or other microbes, we would obviously expect a similar effect depending on the genome size of each microbe.
If there is an upper limit to the number of unique genomes capable of assembly, it doesn’t seem like related fields (i.e. gut microbiome mapping) have documented reaching this despite using whole-genome amplification.



Great article! From what you have read, how might error rates and fidelity of pipolB compare to conventional high-fidelity polymerases, and what implications would that have for downstream sequencing?
Great article, thx for that.
You also mentioned RNA and the necessity of RT, are we sure that all RNA in the sample is completely transcribed into DNA? Random primers work great for standard samples but how does it look for META samples where there should be more RNA in addition to a lot of DNA?
And the second question, can metagenomic samples be prepared in a way that would remove the most represented (and therefore probably already identified) DNA molecules, i.e. to leave only what standard methods have a problem with in the sample