
Note: In this article, I retrace more than a year of my microbiological research, and how it led to the discovery of three new species of viruses (1, 2, 3).
This post was written for citizen scientists and the public. As lengthy as it is, much of it still oversimplifies the nuanced, rapidly-evolving landscape of bacteriophage research.
If you’re a fellow phage researcher, stay tuned for a series of posts where I open-source my protocols and illustrate them in technical detail. Or, feel free to reach me directly at aaryanh@stanford.edu.
Table of Contents
Exposition
A Murder In France
The Beginning of My Adventure
The Search for Samples
Phishing for Phages
Isolating Phages
Sequencing Phage Genomes
The Moment I Discovered a Phage
How to Name Your Phage + Other Side Quests
Aftermath
Acknowledgments
Chapter 0: Exposition
As I slipped on my blue latex gloves in the throbbing California heat, I couldn’t believe that the friendly-looking man I had just emailed yesterday was about to hand me a tube full of sewage, or that the quaint, retrofitted shipping container sitting in front of me was his office.
Before I could reflect on my situation, I was holding a red, plastic bag emblazoned with biohazard logos. On the bike back to lab, I pedaled faster, trying my hardest not to picture the tube warming slowly in my bag, or its unknown contents sloshing around with every pebble and turn. Searching for some moral support, I try craning my neck to the sun, but it only seems to get hotter.
In moments like these, it’s hard to keep the same question from coming to mind: “How did I get here?”
Chapter 1: A Murder in France
Our story begins more than a hundred years ago, in the bustling city of Paris.
Crouched over his workbench, an eccentric scientist by the name of Félix D’Herelle is busy investigating the murder of his bacteria. The evidence in his hands was undeniable—a nutrient plate that should be flooded dysentery colonies, instead wiped clean by an invisible culprit. But who—or what—was to blame?
Like any proper biologist, D’Herelle got to work. After pumping the remains of his bacteria through an ultra-fine Chamberland filter, he found that the liquid that flowed through was just as destructive to his colonies as before. With his elegant experiment, D’Herelle unraveled an important clue: whoever the murderer was, it was far too small to be a bacteria, parasite, or any cell for that matter.
But what was it, exactly?

Months later, still in the thick of his confusion, D’Herelle publishes a landmark paper titled “An invisible, antagonistic microbe of the dysentery bacillus”. Two years later, in 1919, the first patient was cured of dysentery with phage therapy. This was the first documented discovery of a bacteriophage, and perhaps the worst timing share it with the world.
With the isolation of penicillin in the 1930s, excitement in phages waned in favor of cheap, reliable, and scalable antibiotics. In the coming decades, life expectancy skyrocketed, and the global population grew as millions of children survived into adulthood. Humanity had finally liberated itself from the tyrannical reign of bacteria, or so it seemed.
In a strange twist of fate, we found ourselves in much the same condition today. Bacteria are back—and they’re better.
A 2018 study from Addo et al. found that nearly 50% of E. Coli strains isolated from water samples were resistant to one more frontline antibiotics. Roughly speaking, this trend holds across the board, and antimicrobial resistance is projected to take more lives than cancer by 2050.

In retrospect, though, this seems inevitable. Knowing the explosive rates of bacterial replication and mutation, all it takes is a handful of calculation to suggest that bacteria can develop resistance to antibiotics faster they can be designed, tested, put on the shelf.
This is the exact gap that has rekindled phage research around the world. As the natural predators of bacteria, phages satisfy many of the constraints of antibiotics by design. They’re incredibly safe, abundant in nature, and often selective enough to target bacteria with single-strain precision. And unlike antibiotics—which are static molecules that often take years to update— phages have nimbly kept pace with bacterial evolution since the dawn of cellular life.
At a glance, phages seem to embody the perfect antibiotic—that is, assuming we can understand them properly.
Chapter 2: The Beginning of My Journey
Even with its unprecedented comeback, the space of unknowns in phage research remains unfathomably large.
As I write this article, our latest data suggests that humanity has characterized ~15,000 unique phages. For context, we’ve meticulously documented the existence of at least 350,000 species of beetles1.
Save for our fundamental understanding of how phages infect and replicate, the vast majority of phages and hosts haven’t even been described yet. Without a grasp of what phages exist, where they’re metabolized, or how they act, how can we expect to use them in life-or-death situations?
Back when I was a freshman, these were the kinds of open questions that inspired me to join the Bollyky Lab in Stanford’s School of Medicine. This is where I learned just how promising phages are, as well as how enigmatic they still were. After spending nearly two years of investigating our lab’s speciality—phage therapies against Pseudomonas lung infections—I finally had chance to launch a project of my own.
Fortunately, there was an idea that had already been tugging at me. If bacteriophages were the next unexplored frontier in biology, I wanted to help advance it. I wanted to discover a new phage.
Chapter 3: The Search for Samples
One could say Félix left quite the legacy. Fast forward more than century, he’s currently the reason I’m currently riding my bike at full-tilt down Jane Stanford Way.
Which brings me back to my first original question: why was I here?
To understand this, we have to understand what makes viruses special.
In one of his most famous books, the American microbiologist Dickson Despommier once quipped that “successful systems attract parasites”.
Like all viruses, phages are parasites that subsist by entering cells, hijacking their molecular machinery, and turning them into virus-making factories. Since their survival hinges on the presence of an appropriate host, phages tend to congregate around them.
Or, to spin this another way: the simplest way to find a phage is to look for its bacteria of interest.
At the Bollyky Lab, our team focuses on Pseudomonas aeruginosa—an opportunistic bacteria that is already responsible for millions of drug-resistant skin and lung infections around the world.
What makes Pseudomonas particularly interesting are its hideouts. Beyond lingering in hospital rooms, bathroom sinks, and the built human environment, P. Aeruginosa is routinely found in soil, ponds, and wastewater.
With a trail in sight, I decided to spend my first weeks of research as a field biologist.


In 15mL Falcon tubes, I collected stagnated water from Lake Lagunita, scraped soil samples from outside Medical School, and swabbed rooms at Stanford Hospital. Every day meant a new destination, a fascinating person, or another exotic sample to collect.
And that’s how I eventually found myself here, staring up at a weathered, ivy-covered sign that read “Stanford University: Codiga Resource Recovery Center”.
From a series of cold emails and asking around the academic grapevine, I learned that if you wanted wastewater, this was your place, and Bill Li was your guy. It certainly didn’t look like it, but the murky water I had just been gifted held exactly phages I needed.
Which begs the question: how do you get from wastewater to phages?
Chapter 4: Phishing for Phages
Consider zooming into our tube of wastewater.
With enough magnification, it becomes clear that phages aren’t the only residents. If anything, the depths of this liquid resemble a microbial metropolis, where bacteria and fungi tower like skyscrapers, and every sidewalk and street corner teems with viruses and molecules of all kinds.
This quickly presents a dilemma: in such a bustling carnival of microbes, how do we precisely extract the phages we want?
As I soon discovered from closely studying protocols, the short answer is to use the right bait.
In line with most other viruses, phages can only infect bacteria after binding to molecules on their surface. Since this interaction between the surface of a phage and its target is specific, so is the spectrum of bacteria it can infect.
This property—somewhat uncreatively known as host range—is exactly what microbiologists exploit to select phages.
All of this made sense, at least in theory.
But as I stood in our lab and scanned our seemingly endless walls of reagents, I realized that the task on my hands was on a different dimension from the research I had done. By definition, I was looking for something that had never been found before. There were no fifteen-minute guides to follow or reliable results to expect. For the first time, I found myself probing a question that didn’t have clear answers—every experiment was now an act of creativity.
As unnatural as it felt, the only path forward was to try something.
To pull this off in the lab, I began by spinning down our wastewater in a centrifuge. From reading papers like this one, the logic was clear: through its intense rotation, a centrifuge artificially enhances the action of gravity, allowing the components in our samples to spontaneously arrange themselves by density.
Naturally, this means cells and macroscopic particles would aggregate at the bottom of our tube, while viruses and small molecules would form a clear liquid on top called the supernatant. For now, I decided to extract the supernatant and set it aside.
Meanwhile, I wanted to prepare the ideal bait for our phages—in this case, a liquid culture of Pseudomonas bacteria that I brewed up some hours earlier.
What do you think happens to our swimming pool of bacteria when we treat it with our supernatant from earlier?
Let’s play this through in our imagination. If our wastewater contained any trace of Pseudomonas phages, they would immediately infect the bacteria in our culture and replicate at breakneck speed. Any other phages in solution wouldn’t recognize the host, and their numbers would remain stable.
In essence, we get a system where our phages of interest multiply explosively—easily overshadowing the remaining viruses that can’t infect our host.
Of course, carrying this out was slightly more brutal than this summary lets on. Till these experiments, I’d only ever read about phage screening in papers from established labs. Optimizing these experiments from scratch was perhaps the bumpiest stretch of this entire project.

While the first tests took a day to run, honing them into a workable protocol took three weeks of full-time work. It called for a level of testing, critical thinking, and creativity that I never used before.
That said, I still remember how dramatic the results were when this finally worked.
Sitting in a boxy plastic holder were the two defining vials of my experiment. On the left was a negative control—another culture of Pseudomonas that was already taking on a villainous green after a night of incubation. On the right, the same culture treated with our supernatant had already turned a cloudy yellow—the color of trillions of bacteria gushing out their remains at once.
One thing was unmistakable: we had the phages we wanted, and we had a lot of them.
Chapter 5: Isolating Phages
What I said earlier was a lie. Kind of.
If you were careful, maybe you caught it: the method I described lets us begin with any number of phages and end up with only those that infect our bacteria of interest.
But what if our sample contained more than one phage that could infect Pseudomonas?
Before we can say anything meaningful about a phage—whether it’s sequencing, imaging, or anything in between—we need to be sure we’re working with just one kind. Whatever phages our sample harbors, it needs to be as pure as possible.
This presents an even bigger problem than before: how do you separate a population of phages that already infects the same host?
Perhaps unsurprisingly, a quick review of the literature reveals all kinds of solutions, from centrifuging them aggressively to filtering them through sophisticated columns. Having weighed (and tried) many of these options, my weapon of choice soon became a technique called the plaque assay2.
To understand this, let’s return to our vial of phages. On a good run under the right conditions, just a milliliter of our solution can contain 1,000,000,000 (or 10⁹) particles of infectious phages.
If you dropped these phages on a plate of bacteria and waited overnight, you’d end up with what resembles a gaping crater in a field of bacterial proliferation. These structures are known as plaques, and we commonly use them as a proxy to quantify the number of infectious phages in a sample.
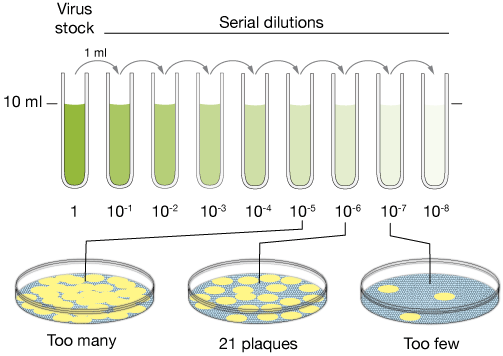
There’s just one issue. Such a potent mix of phages wouldn’t just form a plaque—it would wipe out every trace of bacteria on our plate.
This is useless for same the reason our eyes are useless in blinding light—whether the source is sun or pen laser, the resulting image is undecipherable because it falls far past the threshold of what our eyes can measure. When overwhelmed with phages from all directions, our bacteria suffer the same fate.
The plaque assay overcomes this barrier elegant way: by simply scaling down our population of phages. Instead of completely annihilating bacteria with our concentrated brew of phages, what if we diluted our viruses?
Straight from the vial, our solution is brimming with phages. But with every successive dilution, our phages become fewer and further between, until they finally produce a countable—and therefore, useful—number of plaques.
For instance, if we know that a thousandfold dilution produces ten plaques, we can infer that the original solution would produce ten thousand.
But that wasn’t my goal. While plaque assay lets us count phages, I began wondering if it might also let me separate them. This follows from the same mechanism discussed earlier. With enough rounds of dilution, the hope is that our phages become so sparse that each and every plaque is the doing of a single phage3.
Oftentimes, this process can even be viewed at the macro-scale, since many phages inherently produce plaques of distinct shapes and sizes. If you then dug out just one of these plaques and re-propagated it in bacteria, the resulting batch of phages would turn out far purer than before.
This didn’t work.
For one, it was clear from the first plaque assay that our sample was home to more than one Pseudomonas phage. After enough dilution, what I once thought was a single, unified plaque soon split into a constellation of smaller ones.
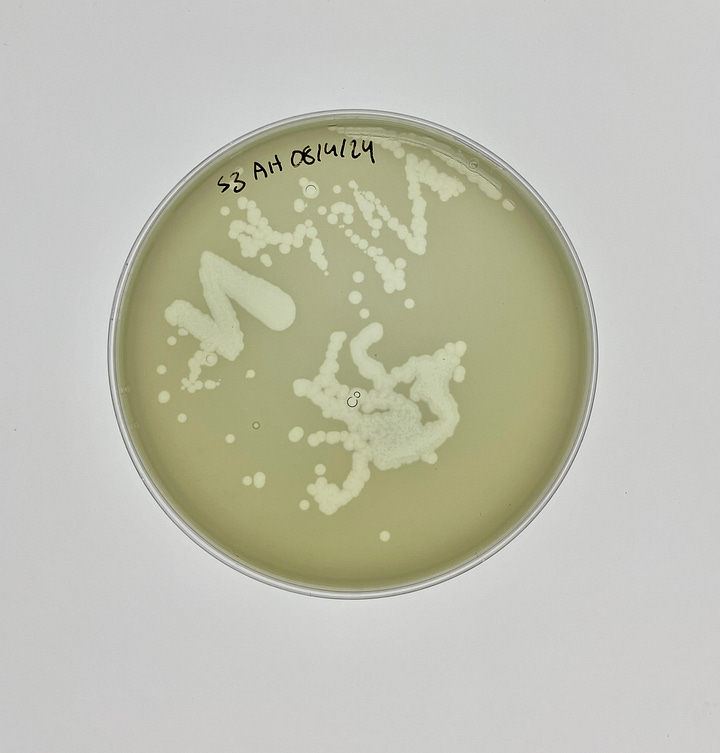

This would have been perfect, if not for a glaring issue. When I extracted the phages from a single plaque to run this test again, it still split into a myriad of unique plaques. The message was clear—one round of purification just wasn’t enough to resolve single phages. Much like before, this was a protocol I ran, reflected on, and reworked for weeks.
In the end, the most reliable technique also happened to be the most painful one. I found that the ideal method to arrive at a pure sample of phage was to run the same plaquing and picking technique five times.
In practice, this meant five straight days of boiling agar in the microwave, holding my breath to pick plaques that were nearly invisible, and tending to my phage cultures like I was the parent of a sleep-deprived newborn.
I’m not sure how felt during those moments, but looking back, I laugh—those solitary nights were perhaps the most important part of my research. It taught me what it meant to pour myself into a question until I found an answer. It taught me how to wrestle with the constant ambiguity of my science. It taught me how to be wrong more times than I can count, and still find the resolve to think of my next attempt.
It was confusing, it was frustrating, it was heart-wrenching at times. And it was the best thing that happened to me as a scientist.
Chapter 6: Sequencing Phage Genomes
On the other side of these experiments, I was looking at six vials that I thought contained distinct phages. Now that their contents were finally pure enough, sequencing was the final step left to understanding their identity.
There was just one problem: our lab didn’t have a sequencer. After Googling for alternatives, asking friends and mentors, and sending a dozen more cold emails, I began feeling like a slice of overly burnt toast.
That same day, fueled by a mixture of desperation and anxiety, something finally snapped. Before I could think through the merits of my strategy, I had already zoomed up two flights of stairs, turned a hallway, and gathered the courage to knock on the door of a lab I’d only heard of before.
The scientists I approached that day belonged to the Gao lab, and they later became instrumental in sequencing my phages. As the summer swept by, many of them became my good friends, and our labs still continue to collaborate long after this project has wrapped up.
With the help of Carlo Armijo, one of Dr. Gao’s PhD students, I learned how to prepare, read, and interpret phage genomes with an Illumina sequencer. And in the months that followed, we had the chance to sequence more phages than I can remember.
If you’ve noticed a common theme by now, you might guess that most of these attempts crashed and burned.

And you would be right. Our very first run, for instance, revealed that our samples still contained fragments of DNA from nearly a dozen different phages. Others produced no reads, or suggested contamination with bacterial DNA. A full two months later, when we thought we had systematically resolved every path to failure, sequencing showed that the phages in our solution already existed.
At this point, I wanted to scream into a pillow.
If this project had any chance of producing novel phages, I would have to start from the beginning. From zero. But as my frustration faded and I reflected on the steps it took to get here, a few explanations began coming into focus.
Perhaps the most glaring issue was my bait. If I was trying to isolate a novel phage—one that no researcher had described before—why was I luring them with the most mainstream strains of Pseudomonas? And if my samples really did contain such a diversity of bacteria, why was I only looking for Pseudomonas phages?
On closer inspection, it wasn’t all too surprising that my experiments had failed.
But I was in far, far too deep to give up now. After taking the weekend off to catch up on lost sleep, I was back, and I was going to set things right.
For one, I snapped on my field gloves and began collecting as many environmental samples as I could. When I wasn’t in the field, I was scouring our storage for the most obscure strains of Pseudomonas I could find. With time, I also expanded my search to E. Coli, which was also supposed to be a common member of the samples I gathered.


This all extended from the same rationale. I figured that the more phages I packed into a single vial, and the more kinds of bacteria that were available to them, the more likely we were to snag an interesting virus.
All counted, it wasn’t until another two months of experiments till I was ready sequence again.
Strangely enough, that might have been the single upshot to all that drudgery—when my sequences finally arrived unscathed, it felt like I’d conquered the universe. Sometimes, I guess the simple pleasures really are the most touching.
Chapter 7: The Moment I Discovered a New Phage
Throughout my runs with Carlo, we relied exclusively on a technique known as sequencing-by-synthesis (SBS).
In this approach, whole phage genomes are chopped up into thousands of overlapping pieces (“reads”) that are then pieced together computationally. After receiving a series of raw reads from our sequencer and assembling them using an algorithm called SPAdes, we could now read the genetic sequence of our phages as clearly as the text in a Google Doc.
But which of these genomes had something interesting to say? Which of these genomes was entirely new?
For a better intuition, we need to first clarify what we mean by a species of virus.
Viruses are notoriously tricky to organize because they aren’t alive. Whereas a species is classically defined as any group of organisms capable of breeding interbreeding, it isn’t clear how this translates to phages that replicate independently.
A more robust definition of a virus comes from the International Committee on the Taxonomy of Viruses (ICTV), which happens to be the global authority on this exact question. Instead of categorizing a virus on its capacity to breed, the ICTV defines a virus as a new species if its genomic sequence is less than 95% similar (or more than 5% different) from its most closely related existing virus4.
As it turns out, there’s a tool for that. When virologists want to quickly understand the genome of an unknown phage, they turn to a program known as the Virus Intergenomic Distance Calculator (VIRIDIC).
If we want to prove the originality of an essay, we use a plagiarism checker. If we want to prove the originality of phage, we use VIRIDIC. In a nutshell, VIRIDIC accepts the genome of any unknown virus and compares it with those of its closest genetic neighbors. If significantly long stretches of the genomes resemble each other, the program determines the degree of similarity, as well as the likelihood that the resemblance was due to chance.
With multiple phages, VIRIDIC outputs a matrix of genetic similarities—much like the results it produced for the phages we sequenced below:
It was hard to miss: from left to right, all the genetic similarities between our phages and their closest viral relatives fell below 95%. According to ICTV’s threshold…I had just discovered at least three new species of viruses.
I refreshed the results, and refreshed them again to make sure. A pause, and the same numbers flashed onscreen.
Nearly a year since I set out to isolate a new virus, I had finally accomplished my mission! After exploring what seemed every possible way to mess up, here I was.
Here I was, but what now?
Chapter 8: How to Name Your Phage + Other Side Quests
Apparently, in the niche corridors of microbiology research, it’s standard practice for the discoverer of a virus to choose its name. Several long walks and conversations later, I eventually made up my mind.
I decided that I’d name my E. Coli phage Cardinal, as a friendly homage to Stanford’s mascot. I’d name the other two Vanta and Luminis—partly because they weren’t already taken, and partly because I thought they sounded cool. (I seriously hope I put more thought into the names of my children).
In the weeks that followed, I found myself surging with inspiration—most of which I channeled into understanding these viruses as much as possible.
For starters, I began teaching myself bioinformatics on walks to lab and between bites of dinner. Eventually, this let me run a program called PHANOTATE to computationally predict the identity and function of all the proteins encoded in phage genomes.
As I refined my skills further, I learned how to render my phages on a proteomic tree, which allowed me to map their relationships based on the similarity of their core proteins, as opposed to the sequences of their nucleic acids.

Armed with this arsenal of tools, I could resolve the classification of my phages in granular detail. For instance, I learned that Cardinal belonged to the family Justusliebigviridae. Vanta and Luminis, on the other hand, belonged to the family Yuaviridae, which hardly contained phages to begin with.
Back in the lab, I propagated our phages, meticulously recounted their numbers, and began screening their activity against a broader library of bacteria. This was around the time I realized these viruses were far more interesting than I imagined.
Partway through my analysis, our lab received a nondescript paper bag from Stanford Hospital. Inside was what seemed like a handful of unremarkable Pseudomonas cultures, with the exception that they were multi-drug resistant isolates from patients with severe lung infections.
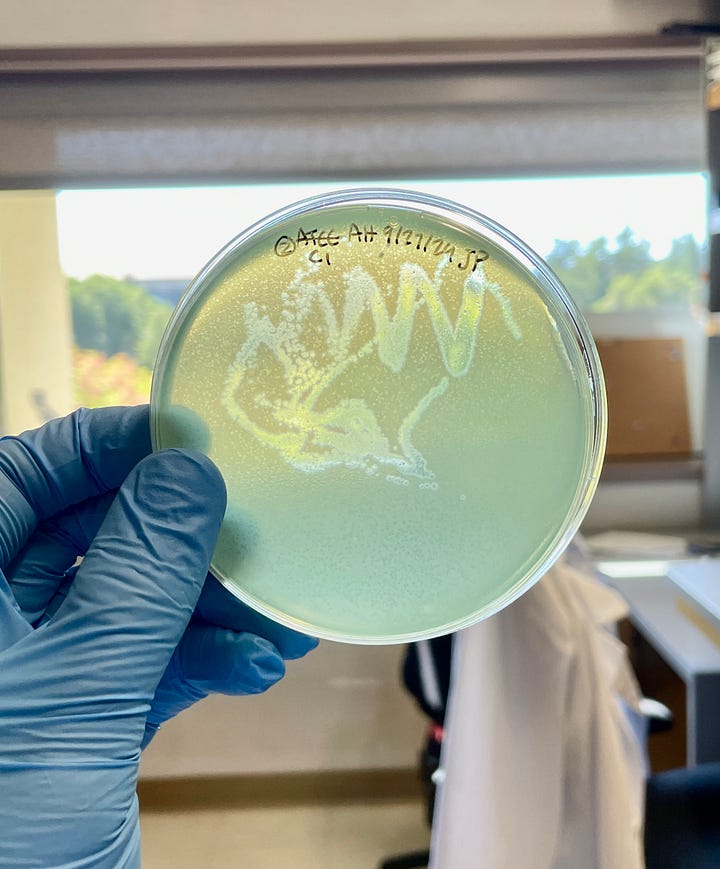
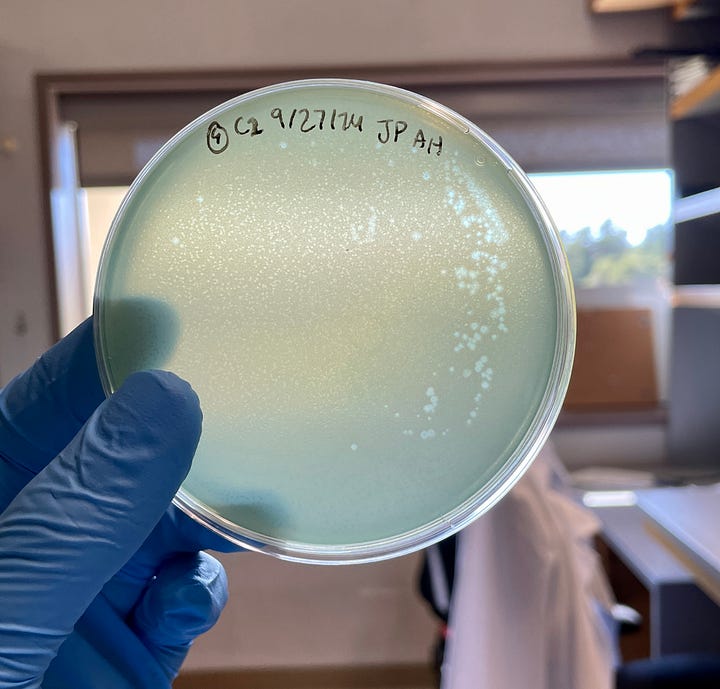
With the approval of my mentor (and a handful more plaque assays), I revealed that both Vanta and Luminis were active against these bacteria, too. It seemed that the phages I isolated weren’t just theoretical curiosities that infected lab bacteria — they formed plaques in clinical strains of Pseudomonas that were already resistant to frontline antibiotics.
While demonstrating the potential of these phages was exciting, it still fell short of the first time I saw them up close.
Much like bioinformatics, I didn’t begin with a shred of experience in imaging viruses. But through some stroke of magic, I did know of Dr. John Perrino, who regularly ran the electron microscope in our basement. And after pestering him with enough emails, he eventually agreed to teach me how to use it.
I still think back to the day everything came together. After spending the morning staining my phages in uranyl acetate, I was now glued to the microscope, holding my breath while scrolling through magnifications as gently as I could manage.
1,200x.
2,000x.
4,500x.
10,000x.
25,000x.
50,000x…
And then—almost as if I’d struck the virological lottery—everything snapped into focus all at once. For the first time, I could see the phages I had isolated—I could see their capsids, their stubby necks, and their long, spindly legs. I could see them in the thousands.



At that moment, it struck me that I might be the first human to ever see this virus. It felt like I was beholding a world I didn’t have permission to see. Mentally, everything went blank except for a single, abiding thought: “Ah, I see now—so this is why people become scientists….”
With all the data I’d collected, I figured it was finally time to make these phages official. After organizing and annotating their genomes, I submitted these phages to NCBI, which currently hosts the world’s largest database of viruses.
Last December, I learned that our phages were live—complete with information on their genome, proteins, and metadata. (If you’re curious, feel free to browse through their entries here — 1, 2, 3).
In the meantime, I compiled the rest of our figures into a report and submitted it to ICTV, which will be reviewing our proposal this May. If these submissions get accepted, all three phages will become official canon recognized by scientists around the world.
For the first time in a long time, all that’s left was to wait and watch.
Chapter 9: Aftermath
This article might have been the most ambitious one I’ve ever written. As I finally type out these concluding words, I decided to return to my favorite writing spot on campus—in this case, a plush, red chair overlooking the entrance of Green Library.
Outside the window, I watch as raindrops begins to drizzle the ground, slowly drenching the world in blue. Almost subconsciously, I catch myself thinking that days like these must have been made for reflection. As the rain quietly patters away, I take a moment to recollect my journey.
Last summer—almost a full year ago—I naïvely set out to discover new viruses. Everything else feels like a blur.
I remember biking frantically around campus as a tube of wastewater sloshed around in my backpack. I remember marathons of plaque assays, and the constant, gnawing fear of seeing a lab ghost alone at night. I remember the confusion of my first day in lab, the rush of our first sequences, the joy of seeing my phages for the first time, and everything in between.
It felt unreal, yet oddly satisfying: one year, three viruses, too many memories to count.
For now, though, I’m in limbo—another transit between what happened and what’s coming. What do I want to do next?
Part of me wants to scale everything. If my shoestring experiments could turn out new viruses in a year, what would happen with ten students and a serious budget? What if I launch Stanford’s first viral discovery initiative? If the future of phages matters so much, I didn’t see any reason why we couldn’t pave the way in the years ahead.
The other part of me wants to get further with the knowledge we already have. I want to learn how phages can be engineered, formulated, and directionally evolved maximize their performance. Since phages are so versatile, I also want to learn how they can be applied outside the clinic—in food production, industrial chemistry, materials science, and biodefense.
Somewhere along the way, I begin to realize just how far I’ve come, and just how little I know, and the immensity of all the questions and ideas that I’m not even aware of yet, and…
I’m suddenly shaken awake by the familiar, high-pitched yelp of my alarm. As my thoughts slowly drift back to my surroundings, I realize that I’m in a library and look like I’ve been hit by a baseball bat. Fumbling for my phone, I remember that I have a long day ahead: bio homework, notes to wrap up for tomorrow, then dinner with friends from art history.
I can’t help but smile a little. Research might be over, but my life is still unfolding.
Till the next mission,
Aaryan Harshith
Chapter 10: Acknowledgments
While this research began as an independent project, it now has a small army of collaborators and friends.
First, I want to thank Dr. Paul Bollyky for taking a chance on me and letting me join his lab as a freshman. I’d also like to thank my direct mentor, Dr. Jessica Sacher, who taught me all the techniques I know and always held me to the highest standards as a scientist.
On that note, this research simply wouldn’t have been possible without the generosity of Bill Li from Stanford’s Resource Recovery Center, and Carlo Armijo from the Gao Lab. I’ll always be grateful for your mentorship and support as this project stopped and started more times than I can remember.
Thank you to Stanford’s Existential Risks Initiative for backing my research last summer, and to all my fellow SERI researchers for teaching me about their work between spoonfuls of ice cream.

And of course, thank you to my family, as well as all my friends on campus and beyond whose presence alone kept me sane. Thank you to Ryan Lee, Grace Bedia, Aspen Singh, Iris Qu, Michelle To, Audrey Jung, Jason Chon, Hemal Arora, Dhevin Nandhyala, Melwin Cheng, Sam Yang, Nicholas Tan, and the many, many more incredible Stanford trees who helped me along my way.
Footnotes
No shade intended. I make this comparison with a deep respect for the importance of beetles, insects, and the study of entomology.
Among the oldest virological techniques that is still employed in modern labs, originally developed in the 1950s by Renato Dulbecco.
Several microbiologists have pointed out that this assumption is problematic, as differentiating between a true plaque and a larger agglomeration of plaques can prove nearly impossible without prior knowledge of a phage’s unique plaque structure.
While this threshold clearly isn’t ideal (especially since genetic differences don’t necessarily imply outward differences in structure or infectivity), it’s held up surprisingly well against the test of time, and is still convenient enough for the ICTV analyse hundreds of species submissions a year.







Phenomenal article Aaryan, I loved it!!! So many surprises even I didn't realize! So cool your phages plaque on those clinical isolates... I've got some more that you can test them on if you want!
This article made me realize I've almost lost the wonder in what we do - it's pretty amazing, and I thank you for writing this as a reminder.
great read aaryan! keep up the great work, love to see it.